Product Studies
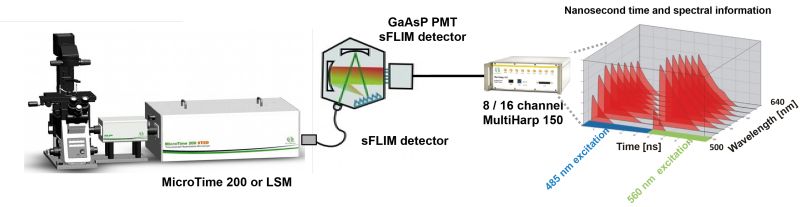
16 channel spectral FLIM detector
Spectrally resolved Fluorescence Lifetime Imaging (sFLIM) for truly parallel multitarget imaging
- Combines lifetime and spectral information for unequivocal identification of a multitude of different fluorescent labels
- Allows for excellent differentiation of multiple fluorescent labels against autofluorescence
- Supports working at high count rates, enabling the acquisition of several sFLIM images per second
Description
Spectral FLIM (sFLIM) is an imaging method that combines spectral resolution and fluorescence lifetime information. Thanks to several improvements, sFLIM can become an extremely fast, versatile and sensitive detection method. One such improvement is the use of a 16 channel spectral FLIM detector. Each of the 16 PMT detector in the array is equipped with a GaAsP cathode, which allow for a detection efficiency of up to 45%. The sFLIM detector array is connected to a MultiHarp 150 featuring 16 parallel TCSPC inputs. Each of these input channel has a very low deadtime of 650 ps, allowing for sFLIM measurements at very high count rates with minimal distortions in the TCSPC histogram and next to no loss of photon counts.
This combination allows for count rates of up to 65 Mcps. Since the fluorescence is simultaneously recorded in several channels, distortions of the fluorescence decay due to detector pulse pile-up are reduced. No photons are discarded, photon loss is kept minimal thanks to the short deadtime of the MultiHarp 1502.
A pattern matching approach has been developed to analyze sFLIM measurements in a fast and reliable manner by fully employing the spectral and lifetime information. Different spectral und lifetime populations can be visualized at a glance by using multiple scatter plots. By defining regions in the scatter plots, reference patterns can be deduced which are then used for linear unmixing of the sFLIM image1. This pattern matching approach is available as a publicly accessible MatLab-based analysis routine (hosted on GitHub). Note that this software is not a regular product from PicoQuant and therefore PicoQuant will not be provideing support for it.
A great application idea for sFIM is measuring multiple fluorescent labels simultaneously such as fluorescent proteins, FRET probes or environment sensitive probes. The fluorescence background due to sample autofluorescence can be also discriminated3.
Application example
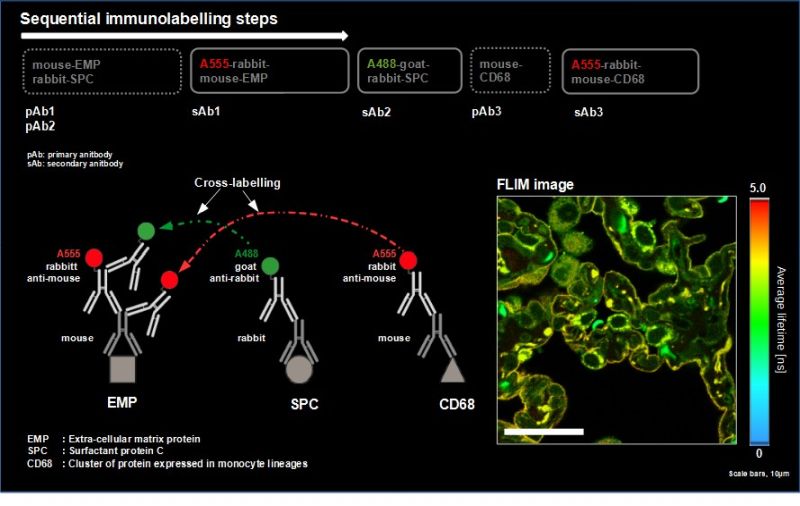
Indirect labeling techniques impose a stringent selection of primary and secondary antibody pairs to avoid false-positive immunolabeling. Due to this, indirect immunofluorescence is regularly limited to two - four antigens or, in the worst cases, particular combinations of target molecule labeling can’t be carried out at all.
In this application example, we present a new method to leverage the ostensible disadvantage of cross-labelling of secondary antibodies by separation of the fluorescence signals by spectral FLIM-FRET. This becomes possible since the undesirable cross-labelling among secondary antibodies leads to the generation of new characteristic FRET emission spectra including a change in the donor lifetime. To demonstrate this, we adapted a sequential labelling protocol and selected appropriate fluorophore pairs on interacting secondary antibodies (e.g., Alexa488 and Alexa546) to achieve strong FRET.
As a model, we labelled the target molecules pan-Cytokeratin, TOM20, and golgin in human lung tissue. The combination of primary and secondary antibodies led to single labelling of TOM20 as well as cross-labelling of pan-Cytokeratin. We used an eight channel spectrally resolved FLIM (sFLIM) detection system and acquired data for all labels excited with two wavelengths operated in Pulsed Interleaved Excitation (PIE) mode.
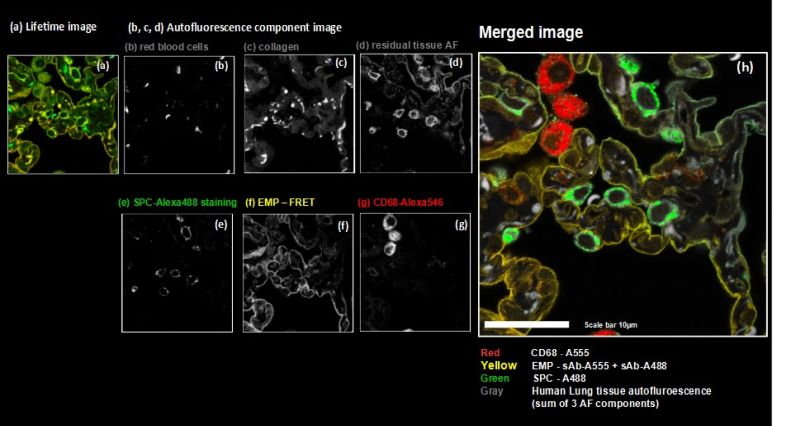
The strong autofluorescence from the human lung tissue – stemming from red blood cells, collagen and residual autofluorescence (AF) – was taken into account by employing three patterns which were derived from an unstained sample. Data analysis was performed using the pattern-matching algorithm1 taking into account emission spectra as well as nanosecond time-resolved fluorescence decays3.
The method enabled us to precisely discriminate between all three target molecules by using the fluorescence generated by just two fluorophore species thanks to their cross-linking and resulting FRET interaction. At the same time, the exogenous fluorescence resulting from three autofluorescent contributions of the human lung tissue could be separated. Consequently, spectral FLIM-FRET together with pattern-matching analysis forms an excellent tool for use in indirect immunofluorescence by overcoming the undesirable effects of secondary antibody cross-labeling and to discriminate against unwanted tissue autofluorescence.
Publications
Thank you for you inquiry.
We have received your message and will contact you as soon as possible.
×