-
Applications
-
Life Science
- rapidFLIMHiRes (Fluorescence Lifetime Imaging) New
- Fluorescence Lifetime Imaging (FLIM)
- Phosphorescence Lifetime Imaging (PLIM)
- Foerster Resonance Energy Transfer (FRET)
- Pulsed Interleaved Excitation (PIE)
- scanning Fluorescence Correlation Spectroscopy (sFCS) New
- Fluorescence Correlation Spectroscopy (FCS)
- Fluorescence Lifetime Correlation Spectroscopy (FLCS)
- Dual-focus Fluorescence Correlation Spectroscopy (2fFCS)
- Stimulated Emission Depletion Microscopy (STED)
- Single Molecule Detection
- Time-resolved Fluorescence
- Fluorescence Anisotropy (Polarization)
- Pattern Matching Analysis
- Two-Photon Excitation (TPE)
- Diffuse Optical Tomography and Imaging
- Singlet Oxygen
- Laser Cutting/Ablation
- Materials Science
- Metrology
- Quantum Optics
-
Life Science
Life Science
rapidFLIM HiRes (Fluorescence Lifetime Imaging) NEW
Combines fast TCSPC data acquisition with high time resolution for reliable, quantitative analysis of FLIM experiments
rapidFLIMHiRes measurements enable the imaging of dynamic processes via fluorescence lifetime imaging (FLIM). This new approach allows for fast FLIM acquisition up to several frames per second for imaging of dynamic processes (e.g., protein interaction, chemical reaction, or ion flux), highly mobile species (e.g., mobility of cell organelles or particles, cell migration), and investigating FRET dynamics. More than 10 frames per second can be acquired, depending on sample brightness and image size.
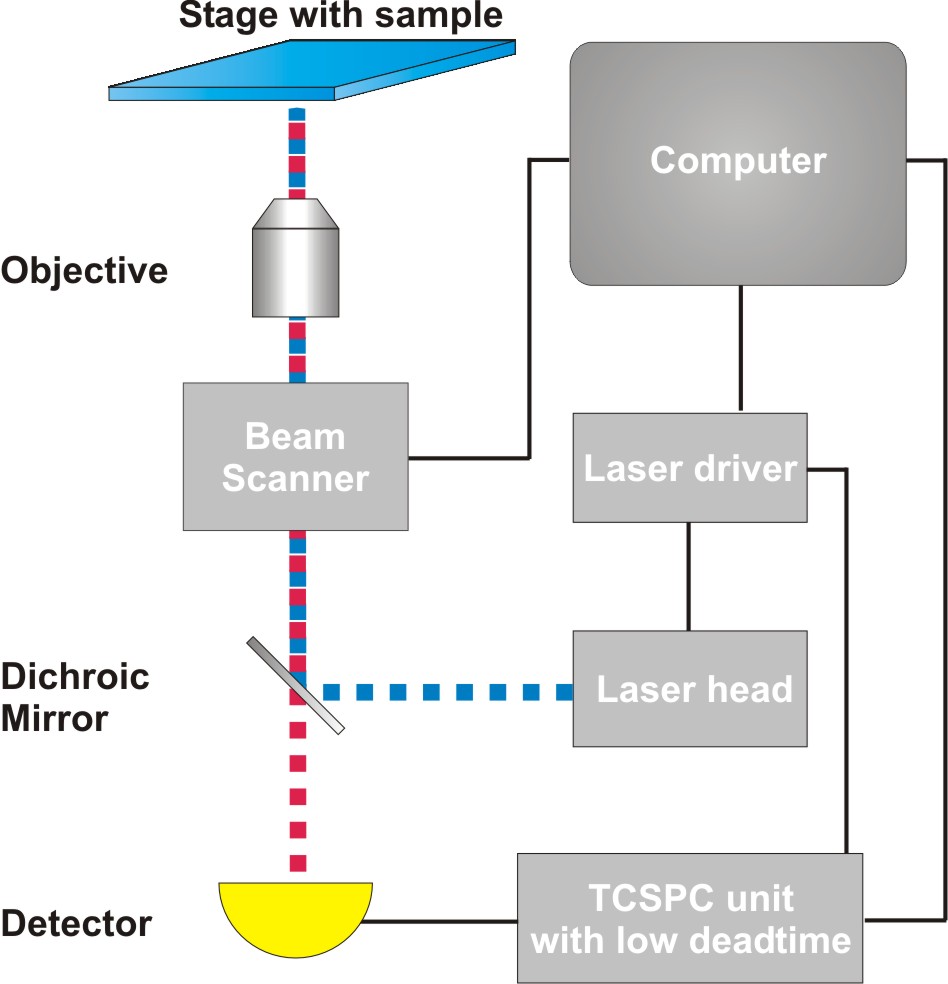 Time-Correlated Single Photon Counting (TCSPC) is used to determine the fluorescence lifetime. In TCSPC, one measures the time between sample excitation by a pulsed laser and the arrival of the emitted photon at the detector very precisely with picosecond accuracy. Classical TCSPC electronics as well as single photon counting detectors have dead times on the order of 80 to 100 ns. Within this time the system is busy with data processing and can not detect any other photon. The dead time is typically longer than the repetition rate of the excitation pulses. Thus, if more than a photon is emitted per excitation cycle, it can not be detected. To avoid this situation that would lead to measurement artefacts, it is necessary to work in conditions in which most of the excitation pulses lead to no emission, i.e. when only receiving one fluorescence photon out of every 50-100 laser pulses. This requirement slows down the FLIM acquisition time.
Time-Correlated Single Photon Counting (TCSPC) is used to determine the fluorescence lifetime. In TCSPC, one measures the time between sample excitation by a pulsed laser and the arrival of the emitted photon at the detector very precisely with picosecond accuracy. Classical TCSPC electronics as well as single photon counting detectors have dead times on the order of 80 to 100 ns. Within this time the system is busy with data processing and can not detect any other photon. The dead time is typically longer than the repetition rate of the excitation pulses. Thus, if more than a photon is emitted per excitation cycle, it can not be detected. To avoid this situation that would lead to measurement artefacts, it is necessary to work in conditions in which most of the excitation pulses lead to no emission, i.e. when only receiving one fluorescence photon out of every 50-100 laser pulses. This requirement slows down the FLIM acquisition time.
In order to acquire a FLIM image with a confocal microscope, one typically scans a pulsed laser beam accross the sample. In case of a fast scanning system, this very often leads to only a few detected photons per pixel, which will not allow proper FLIM analysis. It is therefore necessary to increase the number of detected photons per pixel either by extending the pixel dwell time or by accumulating several frames to sum up the photons. This is another reason why it typically takes several seconds up to half a minute to get a good FLIM image.
The MultiHarp 150 – optimally suited for rapidFLIMHiRes with 5 ps time resolution
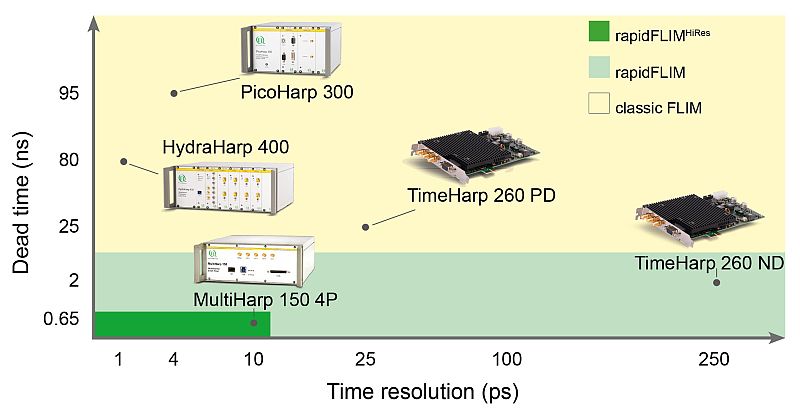 To speed up the data acquisition, a TCSPC unit with a very short dead time like the MultiHarp 150 must be used. The short dead time of less than 650 ps permits to detect several photons within one excitation cycle. The MultiHarp 150 also features a time bin width of 5 ps, that allows determining the lifetimes of commonly used fluorescent probes (usually in the ns range) with outstanding precision. Therfore much higher detector count rates can be processed while achieving highest timing precision. The user no longer has to choose between high time resolution or fast data acquisition.
To speed up the data acquisition, a TCSPC unit with a very short dead time like the MultiHarp 150 must be used. The short dead time of less than 650 ps permits to detect several photons within one excitation cycle. The MultiHarp 150 also features a time bin width of 5 ps, that allows determining the lifetimes of commonly used fluorescent probes (usually in the ns range) with outstanding precision. Therfore much higher detector count rates can be processed while achieving highest timing precision. The user no longer has to choose between high time resolution or fast data acquisition.
Consequently the essential components of a rapidFLIMHiRes set-up are:
- pulsed laser source (diode lasers or multi-photon excitation)
- single photon sensitive detector with low dead time
- dichroic mirror (to separate fluorescence signal from excitation light)
- objective (to focus the excitation light into the sample and collect fluorescence signal)
- TCSPC unit ("stop-watch") with low dead time to measure the time between excitation and fluorescence emission
- fast scanner for imaging
- FLIM acquisition and analysis software
Related technical and application notes:
- Application note: Fluorescence Lifetime Imaging (FLIM) in Confocal Microscopy
- Technical note: Time-Correlated Single Photon Counting (TCSPC)
- Technical note: FLIM: Scanning Speed vs. Excitation Rate
- Application note: Visualize dynamic processes with rapidFLIMHiRes, the ultra fast FLIM imaging method with outstanding time resolution
The rapidFLIMHiRes approach can be integrated into all of PicoQuant’s time-resolved microscopy products
Single Photon Counting Confocal Microscope
Luminosa combines state-of-the-art hardware with cutting edge software to deliver highest quality data while simplifying daily operation. The software includes context-based workflows for FLIM, FRET, and FCS, which allow the users to focus on their samples. Tight integration of hardware and software enable automated routines, e.g., for auto-alignment or laser power calibration, which increase ease of use as well as reproducibility of experiments. Still, an open mode of operation is available for full access to every optomechanical component via software.
LSM Upgrade Kits from PicoQuant add rapidFLIM HiRes to laser scanning microscopes from many major manufactures
Confocal Laser Scanning Microscopes (LSMs) are widely used tools in cell and molecular biology, biochemistry and other related sciences. PicoQuant's LSM Upgrade Kits greatly enhance the capabilities of these microscopes by extension to time-resolved techniques, and thereby providing not only Fluorescence Lifetime Imaging (FLIM) but also Fluorescence Correlation Spectroscopy (FCS) and a wealth of other time resolved techniques. For rapidFLIM, the LSM Upgrade Kit must include a MultiHarp 150 TCSPC unit along with a PMA Hybrid detector possesing a very low effective dead time. Furthermore, a PC with an SSD is required for fast data processing.
Modular, time-resolved confocal fluorescence microscopy platform with rapidFLIMHiRes option
rapidFLIMHiRes experiments can also be carried out with a MicroTime 200 equiped with a FLIMBee galvo scanner, a MultiHarp 150 TCSPC unit along with PMA Hybrid detectors possesing a very low effective dead time. The MicroTime 200 is a time-resolved confocal fluorescence microscope with unique single molecule sensitivity, enabling the realisation of several modalities like FLIM, FCS, STED, anisotropy imaging, anti-bunching measurements in the same microscopic platform.
 Besides bespoke turn-key LSM upgrade kits and the MicroTime 200 microscope, PicoQuant offers the necessary parts to build your own rapidFLIMHiRes set-up from scratch:
Besides bespoke turn-key LSM upgrade kits and the MicroTime 200 microscope, PicoQuant offers the necessary parts to build your own rapidFLIMHiRes set-up from scratch:
- Excitation source with short pulses and high repetition rate
- Laser drivers: Taiko PDL M1, PDL 800-D, PDL 828
- Laser heads: LDH-I Series, LDH Series, LDH-FA Series
- Photon Counting Detector
- TCSPC Electronics
- Scanning device
- Galvo-Scanner
- Analysis software
- Confocal optics
rapidFLIM can be used for several applications such as:
- Following dynamic lifetime processes (e.g., changing interaction states, chemical reaction, highly mobile species)
- Studying fast changes of the fluorescence lifetime caused by e.g., variations of ion concentration or other local parameters such as pH value or oxygen concentration in the fluorophore environment.
- Detecting transient molecular interactions by investigating the FRET dynamics in, e.g., cells
- Monitoring molecular interaction in mobile structures via FRET (e.g., studying mechanisms involved in the mobility of cell organelles, vesicle trafficking, particle movement, or cell migration)
- Perfoming live cell observations such as fast acquisition of FLIM z-stacks, time series, tissue autofluorescence, or dynamics of cell metabolism.
- Using the characteristic autofluorescence for certain tissue types to study, e.g., the dynamics in cell metabolism.
- High throughput FLIM screening
To achieve high detection rates of up to 60-80 Mcounts per second, there must be enough photons coming out of the sample. Low or endogenous expression levels of fluorescent proteins will usually not be enough to work. Currently, GFP seem not to be suitable fluorophore for rapidFLIM, since it shows a lifetime change at higher excitation power.
An alternative to GFP would be the fluorescent protein Venus (see Examples tag), the snap-, halo- and tub-tag labeling technology for site-specific protein modification using organic dyes, or new in vivo STED dyes, such as Silicone Rhodamine from Spirochrome. These dyes are very bright, stable, can be used in vivo, and would overcome the limitations of fluorescent proteins like GFP.
Using fast fluorescence lifetime imaging microscopy to study intracellular Ca2+ signalling upon mechanical stimulation
Intracellular Ca2+ signals induced by mechanical stimulation of HEK-cells loaded with Oregon-Green-Bapta-1. In this video, the cells were imaged before, during, and after mechincal stimulation of the top two cells. The mechanical stimulation leads to an uptake of calcium ions in the cells. This increase in Ca2+ concentration results in a change of the fluorescence lifetime of OGB-1 AM, and the top two cells light up in green for a short time. The rapidFLIMHiRes method enabled imaging a sample area of 128 x 128 pixels with a speed of 5 FLIM frames per second, which makes it possible to quantitatively observe the rise of calcium ion concentration in the cells over a short time period.
Sample details:
- Human Embryonic Kidney (HEK) cells loaded with Oregon-Green-Bapta-1 (OGB-1 AM) kept in artificial cerebrospinal fluid
Set-up:
- 128 x 128 pixel, 3.81 µs pixel dwell time
- 5 frames per second
Sample and data courtesy of Prof. Rose (Institute of Neurobiology, Heinrich Heine University Düsseldorf, Germany)
Using rapidFLIMHiRes imaging to monitor changes in membrane tension
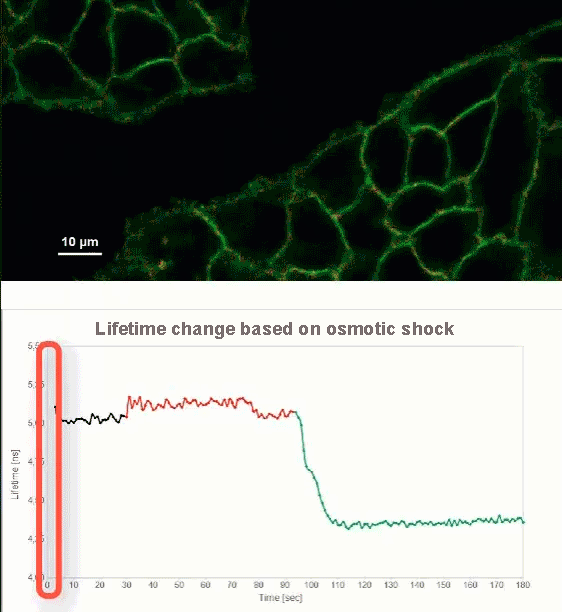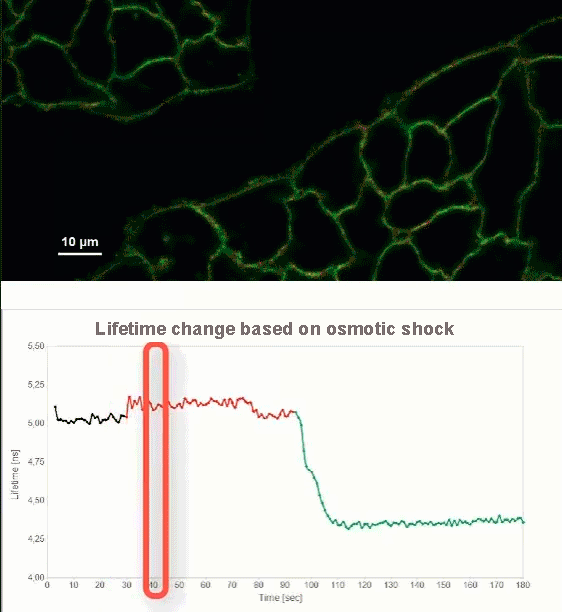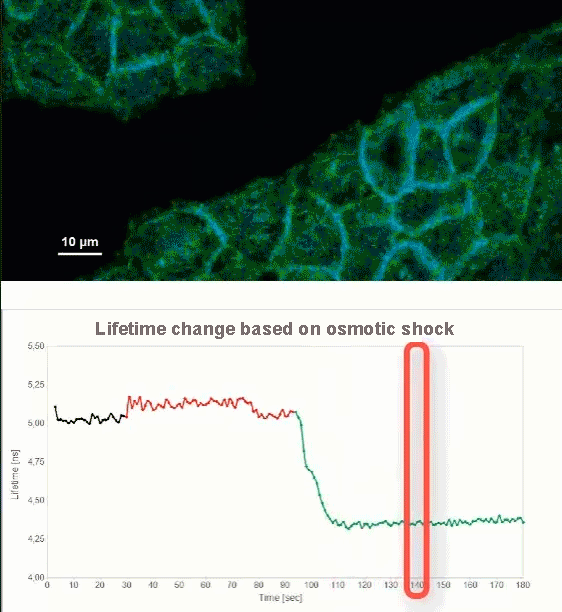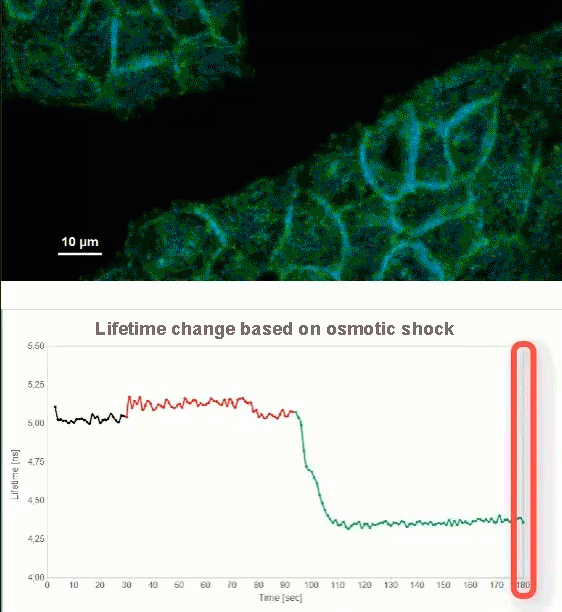
This video shows MDCK cells whose membranes were stained with the fluorescent probe Fliper-TR® (Fluorescent lipid tensor Reporter). This probe displays a significant change in fluorescence lifetime when embedded in a confining environment. If lateral pressure is applied, Fliper-TR® will undergo a planarization which leads to a change in fluorescence lifetime, which allows imaging lipid composition and membrane tension in both live cell and artificial membranes.
Under isoosmotic conditions, Fliper-TR® exhibits a fluorescence lifetime of ca. 5.1 ns. After inducing an hyperosmotic shock, the membrane tension decreases rapidly and a shorter lifetime is observed. Imaging with the rapidFLIMHiRes approach is fast enough so that this lifetime change and by extension the change in membrane tension can be recorded in real time.
Sample details:
- Madin-Darby Canine Kidney (MDCK) cells, membranes stained with Fliper-TR®
Set-up:
- Excitation: 485 nm
- Image size: 512 x 256 pixels
- 1.0 frames per second
Sample courtesy of A. Colom, University of Geneva, Switzerland
Reference: Colom, A., Derivery, E., Soleimanpour, S. et al. A fluorescent membrane tension probe. Nature Chem 10, 1118–1125 (2018). https://doi.org/10.1038/s41557-018-0127-3
Following the motion of unilamellar vesicles with rapid FLIM microscopy
Unilamellar vesicles can be produced in a size regime ranging from giant (GUVs) to large (LUVs), and even to small (SUVs). The flexibility in membrane composition and possibility to introduce specifically labeled lipids increase the importance of unilamellar vesicles for studies in cell biology. Thus turning such vesicles a very powerful model to investigate e.g., membrane domain formation as well as lipid organization in membrane micro-domains.
Until now, acquiring FLIM images took up to several minutes and due to the high mobility of GUV's, imaging them accurately is difficult. Applying the rapidFLIM approach significantly decreases the acquisition time, allowing to record up to several frames per seconds. Thus even highly mobile GUVs can be precisely tracked. In this example, two fluorophore labeled lipids (C6-NBD-PC and N-Rhd-DOPE) were incorporated into GUVs. In non-phase separated GUVs, the lifetime of NBD (7-nitrobenz-2-oxa-1,3-diazol-4-yl) is strongly quenched (down to ~2 ns) due to FRET to the acceptor rhodamine. The video shown here contains 300 frames that were recorded with a frame rate of 5.6 fps.
Sample details:
- GUVs with NBD and Rhodamin labeled lipids (no phase separation): DOPC + 0.5 mol % Palmitoyl-C6-NBD-PC + 0.5 mol % N-Rhd-DOPE
- NBD fused to Palmitoyl-C6-NBD-PC (phosphatidyl choline)
- Rhodamine fused to N-Rhd-DOPE (Di-oleyl-phosphatidyl-ethanolamin)
Set-up:
- Excitation: 485 nm, 40 MHz
- Long pass filter: 488 nm
- 75 x 75 µm, 300 x 300 pixel, 1 µs/pixel
- 300 frames with 5.6 fps
GUV preparation by Ivan Haralampiev, Lab of Molecular Biophysics, Humboldt Universität zu Berlin
Dynamic, in vivo fluorescence lifetime measurements with Venus fused to plant transcription factors
Living mammalian cells expressing two different plant transcription factors (TF) fused to the fluorescent protein Venus were repeatedly scanned with one frame per second (30 µs / px) to follow the protein dynamics within the cytoplasm (type 1 TF, top video) and cell nucleus (type 2 TF, bottom video). Venus shows a stable lifetime and low bleaching at high count rates and thus is suited for rapidFLIM. It was possible to image structures within the nucleus as well as cytoplasmic vesicles with about 1 frame per second. More than 100 photons per pixel could be collected. The bleaching at these high count rates was very low. It could even be minimized by opening the pinhole (sacrifice some resolution) and reducing the laser power. Each of the videos contain 40 frames.
Sample Details:
- Mammalian cells expressing two different plant transcription factors fused to Venus
Set-Up:
- LSM Upgrade Kit at Zeiss LSM 780
- Excitation: 485nm, 32 MHz
- TCSPC: Time Harp 260 NANO
- Detection: > 500 nm
- Number of frames: 40
- Image size: 256 x 256 px
- Pixel size: ~ 300 nm
- Excitation power at objective: 16 µW
- Scan speed: 30 µs / px ~ 1 FPS
- Analysis: SymPhoTime 64
Sample courtesy of Stefanie Weidtkamp-Peters, CAI, University of Düsseldorf, Germany.
Fast fluorescence lifetime imaging enable tracking of phase separated giant unilamellar vesicles
The high flexibility in the lipid composition of giant unilamellar vesicle (GUV) membranes makes them prime candidates for investigating the interactions between lipids and specific micro-domains. For this purpose, we used phase separated GUVs consisting of a lipid ordered (lo) and a lipid disordered (ld) domain. Our rapidFLIM approach allows differentiation of lifetime patterns even in the case of moving vesicles.In phase separated GUVs, the fluorescence decay of C6-NBD-phosphatidylcholine (NBD) contains both a short and a long lifetime component.
The lifetime distribution shows clearly that the longer component (11 ns) is solely found in the lo phase, while the short component (7 ns) can be attributed to the ld phase. Furthermore, the second dye-labeled lipid N-Rhd-DOPE that was added to the GUVs is known to incorporate only into the ld phase. Since the rhodamine dye labeling this lipid can act as a FRET acceptor, the NBD lifetime can be therefor even further reduced in the ld phase. The video shown here contains 200 frames that were recorded with a frame rate of 5.6 fps.
Sample Details:
- GUVs with NBD and Rhodamin (phase separation) : 33 mol % DOPC + 33 mol % stearyl SM (sphingomyelin) + 33 mol % Cholesterol + 0.5 mol % Palmitoyl-C6-NBD-PC + 0.5 mol % N-Rhd-DOPE
- Rhodamine fused to N-Rhd-DOPE (Di-oleyl-phosphatidyl-ethanolamin)
- → disordered phase (ld): 7ns
- → ordered phase (lo): 11 ns
Set-Up:
- Excitation: 485 nm, 40 MHz
- Filter: long pass 488 nm
- 75x75 µm, 300x300 pixel, 1 µs / pixel
- 200 frames recorded with 5.6 fps
GUV preparation by Ivan Haralampiev, Lab of Molecular Biophysics, Humboldt Universität zu Berlin
Dynamic recording of fluorescence intensity images allows studying bead diffusion behavior
The diffusion behavior of two kinds of dye-labeled beads, monitored via rapidFLIM, is shown. The large beads with a size of 3.42 µm are labeled with Nile Red showing a fluorescence lifetime of 3.3 ns. The smaller bead population (average diameter of 2.07 µm) contains the dye Dragon Green featuring a fluorescence lifetime of 4.0 ns. The two species can be distinguished based on their lifetime difference of about 700 ps. The beads form a random structure at the edge of the water droplet, which is subsequently disrupted by addition of another water droplet. The diffusion processes lead to the reassembly of a similar structure over time. The movie was acquired with 8 µs pixel dwell time corresponding to about 3 frames per second. In one frame maximum 300 photons per pixel were detected.
Sample Details:
- Nile Red: Lifetime 3.3 ns, bead size 3.42 µm
- Dragon Green: Lifetime 4.0 ns, bead size 2.07 µm
Set-Up:
- LSM Upgrade Kit on Olympus FluoView FV1000
- Excitation: 485 nm, 20 MHz
- TCSPC unit: Time Harp 260 NANO
- Detector: PMA Hybrid, > 500 nm
- Image size: 128 x 128 px
- Scan speed: 8 µs / px ~ 3 FPS
- Max. 300 photons per pixel
- Analysis: SymPhoTime 64
Applying pattern matching analysis to lifetime images for studying bead diffusion dynamics
The diffusion behavior of two types of dye-labeled beads was monitored via rapidFLIM, as above. Doubling the image acquisition speed by reducing the pixel dwell time towards 4 µs (about 6 frames per second) results in more noise in the FastFLIM image leading to a lower FLIM contrast, as can be seen in the top video.
The lifetime contrast can be enhanced by applying the pattern matching approach. For each species, a lifetime pattern was generated (shown below: green curve = Dragon Green, red curve = Nile Red) and the recorded FLIM data was analyzed, resulting in the second video with improved lifetime contrast.
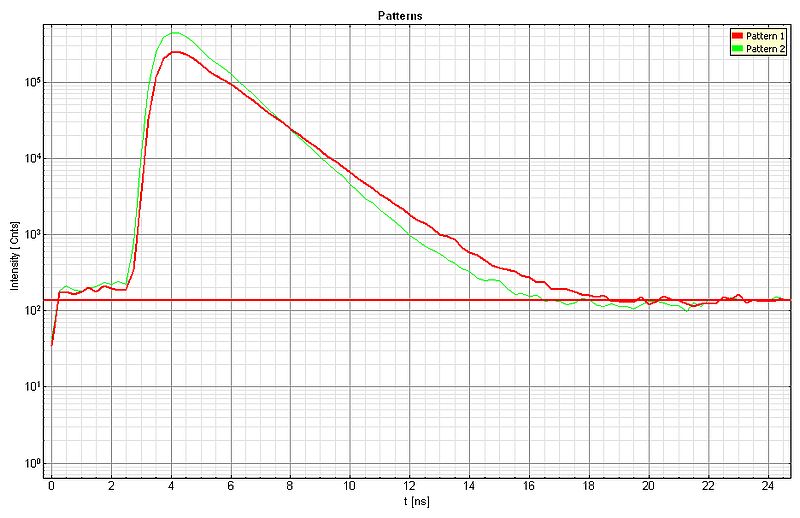
Sample Details:
- Nile Red: Lifetime 3.3 ns, bead size 3.42 µm
- Dragon Green: Lifetime 4.0 ns, bead size 2.07 µm
- LSM Upgrade Kit on Olympus FluoView FV1000
- Excitation: 485 nm, 20 MHz
- TCSPC unit: Time Harp 260 NANO
- Detector: PMA Hybrid, > 500 nm
- Image size: 128 x 128 px
- Scan speed: 4 µs / px ~ 6 FPS
- Max. 300 photons per pixel
- Analysis: SymPhoTime 64
Latest 10 publications related to FLIM
The following list is an extract of 10 recent publications from our bibliography that either bear reference or are releated to this application and our products in some way. Do you miss your publication? If yes, we will be happy to include it in our bibliography. Please send an e-mail to info@picoquant.com containing the appropriate citation. Thank you very much in advance for your kind co-operation.

 Get in touch
Get in touch
 Luminosa
Luminosa LSM Upgrade Kits
LSM Upgrade Kits